Note: This vignette uses fake data - it is for illustrative purposes only and should not be used to inform decision making. This vignette outlines the workflow for developing utility mapping models using longitudinal data. The workflow for developing utility mapping models is broadly similar, with some minor modifications. An example of developing models using cross-sectional data is available at https://doi.org/10.5281/zenodo.8098595 .
Motivation
Health services do not typically collect health utility data from their clients, which makes it more difficult to place an economic values on outcomes attained in these services. One strategy for addressing this gap is to use data from similar samples of patients that contain both health utility and the types of outcome measures that are collected in clinical services. The TTU package provides a toolkit for conducting and reporting a utility mapping (or Transfer to Utility) study.
Implementation
The TTU package contains modules of the ready4 youth mental health economic model that combine and extend model modules for:
- labeling, validating and summarising youth mental health datasets (from the youthvars package);
- scoring health utility (from the scorz package);
- specifying and testing statistical models (from the specific package);
- generating reproducible analysis reports (from the ready4show package); and
- sharing data via online data repositories (from the ready4use package).
Additionally, TTU relies on two RMarkdown programs:
- ttu_mdl_ctlg: Generate a Template Utility Mapping (Transfer to Utility) Model Catalogue (https://doi.org/10.5281/zenodo.5936870)
- ttu_lng_ss: Create a Draft Scientific Manuscript For A Utility Mapping Study (https://doi.org/10.5281/zenodo.5976987)
Outputs generated by the TTU package are designed to be compatible with health economic models developed with the ready4 framework).
Workflow
Background and citation
The following workflow illustrates (using fake data) the same steps we used in a real world study, a summary of which is available at https://doi.org/10.1101/2021.07.07.21260129). Citation information for that study is:
@article {Hamilton2021.07.07.21260129,
author = {Hamilton, Matthew P and Gao, Caroline X and Filia, Kate M and Menssink, Jana M and Sharmin, Sonia and Telford, Nic and Herrman, Helen and Hickie, Ian B and Mihalopoulos, Cathrine and Rickwood, Debra J and McGorry, Patrick D and Cotton, Sue M},
title = {Predicting Quality Adjusted Life Years in young people attending primary mental health services},
elocation-id = {2021.07.07.21260129},
year = {2021},
doi = {10.1101/2021.07.07.21260129},
publisher = {Cold Spring Harbor Laboratory Press},
URL = {https://www.medrxiv.org/content/early/2021/07/12/2021.07.07.21260129},
eprint = {https://www.medrxiv.org/content/early/2021/07/12/2021.07.07.21260129.full.pdf},
journal = {medRxiv}
}The program applied in that study, which this workflow closely resembles is available at https://doi.org/10.5281/zenodo.6116077 and can be cited as follows:
@software{hamilton_matthew_2022_6212704,
author = {Hamilton, Matthew and
Gao, Caroline},
title = {{Complete study program to reproduce all steps from
data ingest through to results dissemination for a
study to map mental health measures to AQoL-6D
health utility}},
month = feb,
year = 2022,
note = {{Matthew Hamilton and Caroline Gao (2022).
Complete study program to reproduce all steps from
data ingest through to results dissemination for a
study to map mental health measures to AQoL-6D
health utility. Zenodo.
https://doi.org/10.5281/zenodo.6116077. Version
0.0.9.3}},
publisher = {Zenodo},
version = {0.0.9.3},
doi = {10.5281/zenodo.6212704},
url = {https://doi.org/10.5281/zenodo.6212704}
}Set consent policy
By default, methods associated with TTU modules will request your
consent before writing files to your machine. This is the safest option.
However, as there are many files that need to be written locally for
this program to execute, you can overwrite this default by supplying the
value “Y” to methods with a consent_1L_chr argument.
consent_1L_chr <- "" # Default value - asks for consent prior to writing each file.Add dataset metadata
We use the Ready4useDyad and Ready4useRepos modules to retrieve and ingest and to then pair a dataset and its data dictionary.
A <- Ready4useDyad(ds_tb = Ready4useRepos(dv_nm_1L_chr = "fakes", dv_ds_nm_1L_chr = "https://doi.org/10.7910/DVN/HJXYKQ", dv_server_1L_chr = "dataverse.harvard.edu") %>%
ingest(fls_to_ingest_chr = c("ymh_clinical_tb"), metadata_1L_lgl = F) %>% youthvars::transform_raw_ds_for_analysis(),
dictionary_r3 = Ready4useRepos(dv_nm_1L_chr = "TTU", dv_ds_nm_1L_chr = "https://doi.org/10.7910/DVN/DKDIB0", dv_server_1L_chr = "dataverse.harvard.edu") %>%
ingest(fls_to_ingest_chr = c("dictionary_r3"), metadata_1L_lgl = F)) %>%
renew(type_1L_chr = "label")We use the YouthvarsSeries module to supply metadata about our longitudinal dataset vignette.
A <- YouthvarsSeries(a_Ready4useDyad = A, id_var_nm_1L_chr = "fkClientID", timepoint_var_nm_1L_chr = "round",
timepoint_vals_chr = levels(procureSlot(A, "ds_tb")$round))Score health utility
We next use the ScorzAqol6Adol module to score adolescent AQoL-6D health utility.
A <- TTUProject(a_ScorzProfile = ScorzAqol6Adol(a_YouthvarsProfile = A))
A <- renew(A, what_1L_chr = "utility") ## Joining with `by = join_by(fkClientID, match_var_chr)`Evaluate candidate models
Over the next few steps we will use modules from the specific package to specify and assess a number of candidate utility mapping models.
Specify modelling parameters
We begin by specifying the parameters we will use in our modelling
project. The initial step is to ensure the fields in A for
storing parameter values are internally consistent with the data we have
entered in the previous steps.
A <- renew(A, what_1L_chr = "parameters")We next ingest a lookup table of metadata about the variables we plan to explore as candidate predictors. In this case, we are sourcing the lookup table from an online data repository.
A <- renew(A, "use_renew_mthd", fl_nm_1L_chr = "predictors_r3", type_1L_chr = "predictors_lup",
y_Ready4useRepos = Ready4useRepos(dv_nm_1L_chr = "TTU", dv_ds_nm_1L_chr = "https://doi.org/10.7910/DVN/DKDIB0",
dv_server_1L_chr = "dataverse.harvard.edu"),
what_1L_chr = "parameters")We can inspect the metadata on candidate predictors that we have just ingested.
We add additional metadata about variables in our dataset that will be used in exploratory modelling.
A <- renew(A, c(0.03,1), type_1L_chr = "range", what_1L_chr = "parameters") %>%
renew(c("BADS","GAD7", "K6", "OASIS", "PHQ9", "SCARED"),
type_1L_chr = "predictors_vars", what_1L_chr = "parameters") %>%
renew(c("d_sex_birth_s", "d_age", "d_sexual_ori_s", "d_studying_working", "c_p_diag_s", "c_clinical_staging_s", "SOFAS"),
type_1L_chr = "covariates", what_1L_chr = "parameters") %>%
renew(c("d_age","Gender","d_relation_s", "d_sexual_ori_s" ,"Region", "d_studying_working", "c_p_diag_s", "c_clinical_staging_s","SOFAS"),
type_1L_chr = "descriptives", what_1L_chr = "parameters") %>%
renew("d_interview_date", type_1L_chr = "temporal", what_1L_chr = "parameters")We record that the data we are working with is fake (this step can be skipped if working with real data).
A <- renew(A, T, type_1L_chr = "is_fake", what_1L_chr = "parameters")We update A for internal consistency with the values we
have previously supplied and create a local workspace to which output
files will be written.
We now generate tables and charts that describe our dataset. These are saved in a sub-directory of our output data directory, and are available for download. One of the plots is also reproduced here.
A <- author(A, consent_1L_chr = consent_1L_chr, digits_1L_int = 3L, what_1L_chr = "descriptives")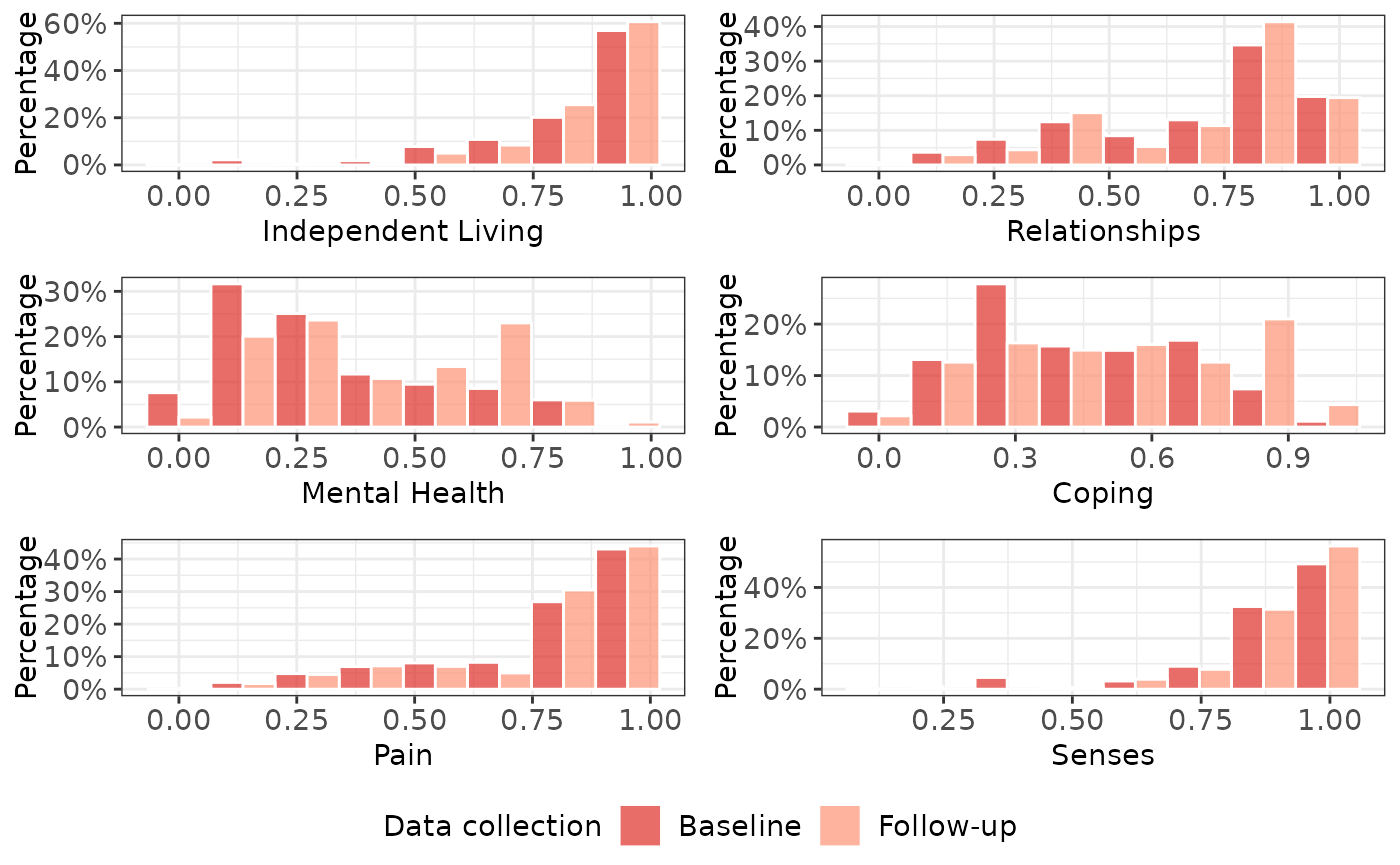
We next compare the performance of different model types. We perform
this step using the investigate method. This is the first
of several times that we use this method. Each time the method is called
A is updated to that the next time the method is called, a
different algorithm will be used. The sequence of calls to
investigate is therefore important (it should be in the
same order as outlined in this example and you should not attempt to
repeat a call to investigate to redo a prior step).
A <- investigate(A, consent_1L_chr = consent_1L_chr, depnt_var_max_val_1L_dbl = 0.9999, session_ls = sessionInfo())The outputs of the previous command are saved into a sub-directory of our output directory. An example of this output is available for download). Once we inspect this output, we can then specify the preferred model types to use from this point onwards.
Next we assess multiple versions of our preferred model type - one single predictor model for each of our candidate predictors and the same models with candidate covariates added.
A <- investigate(A, consent_1L_chr = consent_1L_chr)The previous step saved output into a sub-directory of our output directory. Example output is available for download: (single predictor comparisons) and multivariate model comparisons. After reviewing this output, we can specify the covariates we wish to add to the models we will assess from this point forward.
A <- renew(A, "SOFAS", type_1L_chr = "covariates", what_1L_chr = "results")We can now assess the multivariate models.
A <- investigate(A, consent_1L_chr = consent_1L_chr)As a result of the previous step, more model objects and plot files have been saved to a sub-directory of our output directory. Examples of this output are available for download here and here. Once we inspect this output we can reformulate the models we finalised in the previous step so that they are suitable for modelling longitudinal change. For our primary analysis, we use a mixed model formulation of the models that we previously selected. A series of large model files are written to the local output data directory.
A <- investigate(A, consent_1L_chr = consent_1L_chr)For our secondary analyses, we specify alternative combinations of predictors and covariates.
A <- investigate(A, consent_1L_chr = consent_1L_chr,
scndry_anlys_params_ls = make_scndry_anlys_params(candidate_predrs_chr = c("SOFAS"),
candidate_covar_nms_chr = c("d_sex_birth_s", "d_age", "d_sexual_ori_s", "d_studying_working"),
prefd_covars_chr = NA_character_) %>%
make_scndry_anlys_params(candidate_predrs_chr = c("SCARED","OASIS","GAD7"),
candidate_covar_nms_chr = c("PHQ9", "SOFAS", "d_sex_birth_s", "d_age", "d_sexual_ori_s", "d_studying_working"),
prefd_covars_chr = "PHQ9"))Report findings
Create shareable models
The model objects created and saved in our working directory by the preceding steps are not suitable for public dissemination. They are both too large in file size and, more importantly, include copies of our source dataset. We can overcome these limitations by creating shareable versions of the models. Two types of shareable version are created - copies of the original model objects in which fake data overwrites the original source data and summary tables of model coefficients.
A <- author(A, consent_1L_chr = consent_1L_chr, what_1L_chr = "models")Specify study reporting metadata
We update A so that we can begin use it to render and
share reports.
A <- renew(A, what_1L_chr = "reporting")We add metadata relevant to the reports that we will be generating to these fields. Note that the data we supply to the Ready4useRepos object below must relate to a repository to which we have write permissions (otherwise subsequent steps will fail).
A <- renew(A, ready4show::authors_tb, type_1L_chr = "authors", what_1L_chr = "reporting") %>%
renew(ready4show::institutes_tb, type_1L_chr = "institutes", what_1L_chr = "reporting") %>%
renew(c(3L,3L), type_1L_chr = "digits", what_1L_chr = "reporting") %>%
renew(c("PDF","PDF"), type_1L_chr = "formats", what_1L_chr = "reporting") %>%
renew("A hypothetical utility mapping study using fake data", type_1L_chr = "title", what_1L_chr = "reporting") %>%
renew(renew(ready4show_correspondences(), old_nms_chr = c("PHQ9", "GAD7"), new_nms_chr = c("PHQ-9", "GAD-7")), type_1L_chr = "changes", what_1L_chr = "reporting") %>%
renew(Ready4useRepos(dv_nm_1L_chr = "fakes", dv_ds_nm_1L_chr = "https://doi.org/10.7910/DVN/D74QMP", dv_server_1L_chr = "dataverse.harvard.edu"), type_1L_chr = "repos", what_1L_chr = "reporting") Author model catalogues
We download a program for generating a catalogue of models and use it to summarising the models created under each study analysis (one primary and two secondary). The catalogues are saved locally.
A <- author(A, consent_1L_chr = consent_1L_chr, download_tmpl_1L_lgl = T, what_1L_chr = "catalogue")Author manuscript
We add some content about the manuscript we wish to author.
A <- renew(A, "Quality Adjusted Life Years (QALYs) are often used in economic evaluations, yet utility weights for deriving them are rarely directly measured in mental health services.",
type_1L_chr = "background", what_1L_chr = "reporting") %>%
renew("None declared", type_1L_chr = "conflicts", what_1L_chr = "reporting") %>%
renew("Nothing should be concluded from this study as it is purely hypothetical.", type_1L_chr = "conclusion", what_1L_chr = "reporting") %>%
renew("The study was reviewed and granted approval by no-one." , type_1L_chr = "ethics", what_1L_chr = "reporting") %>%
renew("The study was funded by no-one.", type_1L_chr = "funding", what_1L_chr = "reporting") %>%
renew("three months", type_1L_chr = "interval", what_1L_chr = "reporting") %>%
renew(c("anxiety", "AQoL","depression", "psychological distress", "QALYs", "utility mapping"), type_1L_chr = "keywords", what_1L_chr = "reporting") %>%
renew("The study sample is fake data.", type_1L_chr = "sample", what_1L_chr = "reporting") We create a brief summary of results that can be interpreted by the program that authors the manuscript.
A <- renew(A, c("AQoL-6D", "Adolescent AQoL Six Dimension"), type_1L_chr = "naming", what_1L_chr = "reporting")
A <- renew(A, "use_renew_mthd", type_1L_chr = "abstract", what_1L_chr = "reporting")We create and save the plots that will be used in the manuscript.
A <- author(A, consent_1L_chr = consent_1L_chr, what_1L_chr = "plots")We download a program for generating a template manuscript and run it to author a first draft of the manuscript.
A <- author(A, consent_1L_chr = consent_1L_chr, download_tmpl_1L_lgl = T, what_1L_chr = "manuscript")We can copy the RMarkdown files that created the template manuscript to a new directory (called “Manuscript_Submission”) so that we can then manually edit those files to produce a manuscript that we can submit for publication.
A <- author(A, consent_1L_chr = consent_1L_chr, type_1L_chr = "copy", what_1L_chr = "manuscript")At this point in the workflow, additional steps are required to adapt / author the manuscript that will be submitted for publication. However, in this example we are going to skip that step and keep working with the unedited template manuscript. If we had a finalised manuscript authoring program stored online, we could now specify the repository from which the program can be retrieved.
# Not run
# A <- renew(A, c("URL of GitHub repository with", "Program version number"), type_1L_chr = "template-manuscript", what_1L_chr = "reporting")We can now configure the output to be generated by the manuscript authoring program. The below commands will specify a Microsoft Word format manuscript and a PDF technical appendix. Unlike the template manuscript, the figures and tables will be positioned after (and not within) the main body of the manuscript. Note that the Word version of the manuscript generated by these values will require some minor formatting edits (principally to the display of tables and numbering of sections).
A <- renew(A, F, type_1L_chr = "figures-body", what_1L_chr = "reporting") %>%
renew(F, type_1L_chr = "tables-body", what_1L_chr = "reporting") %>%
renew(c("Word","PDF"), type_1L_chr = "formats", what_1L_chr = "reporting")Once any edits to the RMarkdown files for creating the submission
manuscript have been finalised, we can run the following command to
author the manuscript. If we are using a custom manuscript authoring
program downloaded from an online repository the
download_tmpl_1L_lgl argument will need to be set to
T.
A <- author(A, consent_1L_chr = consent_1L_chr, download_tmpl_1L_lgl = F, type_1L_chr="submission", what_1L_chr = "manuscript")We can now generate the Supplementary Information for the submission manuscript.
A <- author(A, consent_1L_chr = consent_1L_chr, supplement_fl_nm_1L_chr = "TA_PDF", type_1L_chr="submission", what_1L_chr = "supplement")Share outputs
We can now share non-confidential elements (ie no copies of
individual records) of the outputs that we have created via our study
online repository. To run this step you will need write permissions to
the online repository. In the below step we are sharing model
catalogues, details of the utility instrument, the shareable mapping
models (designed to be used in conjunction with the youthu
package), our manuscript files and our supplementary information. In
most real world studies the manuscript would not be shared via an online
repository - the what_chr argument would need to be
ammended to reflect this.
A <- share(A, types_chr = c("auto", "submission"), what_chr = c("catalogue", "instrument" ,"manuscript", "models", "supplement"))The dataset we created in the previous step is viewable here: https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/D74QMP
Tidy workspace
The preceding steps saved multiple objects (mostly R model objects) that have embedded within them copies of the source dataset. To protect the confidentiality of these records we can now purge all such copies from our output data directory.
A <- author(A, what_1L_chr = "purge")